Quickstart#
import pytometry as pm
import readfcs
import anndata
Read fcs file example from the readfcs package.
path_data = readfcs.datasets.example()
adata = pm.io.read_fcs(path_data)
/home/runner/work/pytometry/pytometry/.nox/build-3-9/lib/python3.9/site-packages/fcsparser/api.py:440: UserWarning: The default channel names (defined by the $PnS parameter in the FCS file) were not unique. To avoid problems in downstream analysis, the channel names have been switched to the alternate channel names defined in the FCS file. To avoid seeing this warning message, explicitly instruct the FCS parser to use the alternate channel names by specifying the channel_naming parameter.
warnings.warn(msg)
assert isinstance(adata, anndata._core.anndata.AnnData)
pm.pp.split_signal(adata, var_key="channel")
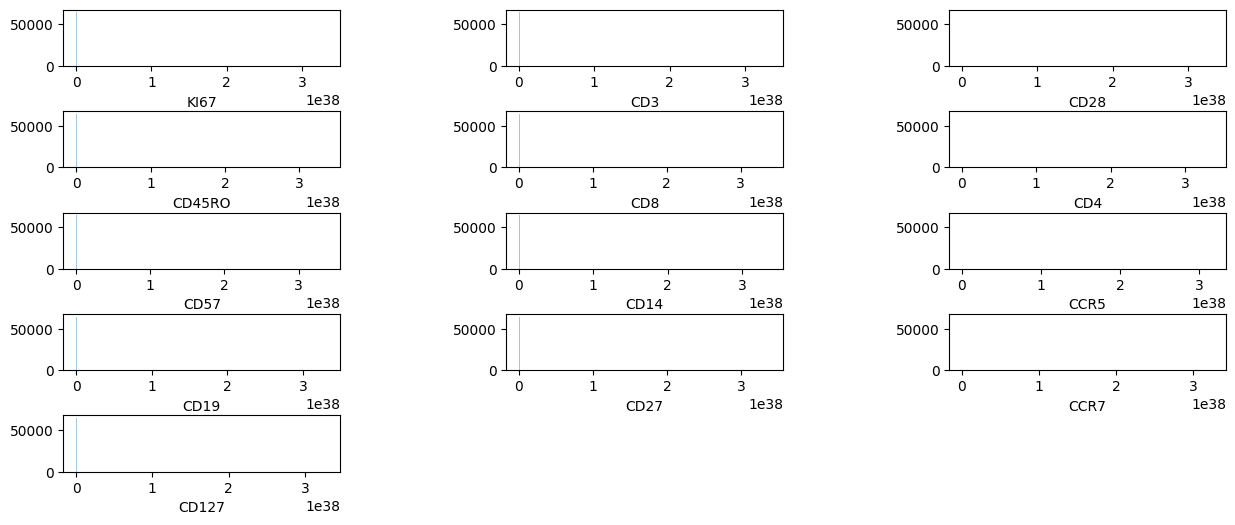
pm.pl.plotdata(adata)
/home/runner/work/pytometry/pytometry/.nox/build-3-9/lib/python3.9/site-packages/pytometry/plotting/_histogram.py:83: UserWarning:
`distplot` is a deprecated function and will be removed in seaborn v0.14.0.
Please adapt your code to use either `displot` (a figure-level function with
similar flexibility) or `histplot` (an axes-level function for histograms).
For a guide to updating your code to use the new functions, please see
https://gist.github.com/mwaskom/de44147ed2974457ad6372750bbe5751
sns.distplot(
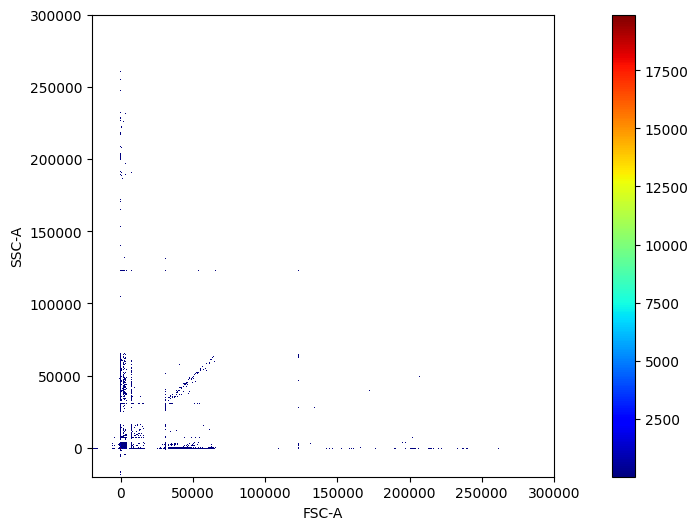
pm.pl.scatter_density(adata)
pm.pp.compensate(adata)
5499 NaN values found after compensation. Please adjust compensation matrix.
adata_arcsinh = pm.tl.normalize_arcsinh(adata, cofactor=150, inplace=False)
adata_biexp = pm.tl.normalize_biExp(adata, inplace=False)
adata_logicle = pm.tl.normalize_logicle(adata, inplace=False)
/home/runner/work/pytometry/pytometry/.nox/build-3-9/lib/python3.9/site-packages/pytometry/tools/_normalization.py:166: RuntimeWarning: invalid value encountered in double_scalars
y = (ae2bx + p["f"]) - (ce2mdx + value)
Save data to HDF5 file format.
adata.write("example.h5ad")